Future of Life Sciences: The convergence of life and technology
Melanie Swan, MS Futures Group, m@melanieswan.com, 415-505-4426
April 3, 2009
Slides

In the next few decades, human capability could be surpassed by machine capability. However, life and technology are not disparate streams but rather convergent as high-impact research findings show in synthetic biology, DNA nanotechnology, nanomedicine, neuroimaging, whole brain simulation and longevity.
The agenda is first an introduction to the notion of the convergence of life and technology, second, a review of recent high-impact research and third, a look at future implications.

Full human brain neural simulation estimated: 2018
Thinking about the future of life sciences, a central issue quickly comes up regarding intelligence and the future of intelligence. For the future of intelligence, there are two main contenders, the human and the machine (here in the blue inlay boxes).
The fastest supercomputer is the IBM Roadrunner, operating at 1 petaflop, 1015 flops, or 1,000 trillion Instructions Per Second and has an 80 TebiByte memory. The average human, on the other hand, runs at an estimated 20,000 trillion Instructions Per Second and has a 1,000 terrabyte memory. So right now our fastest supercomputer is just 5% the capacity of the whole human brain, …but supercomputers are firmly on Moore’s law curves as this chart demonstrates where the human brain is not.
The next supercomputer node, one more order of magnitude, 1016 flops, is expected in 2011 with the Pleiades, Blue Waters and Japanese RIKEN systems. 1016 flops would possibly allow human brain functional simulation. But the big leap could be three more orders of magnitude to 1019 flops, into the exaflop computing era, expected in 2018, allowing full human brain neural simulation. This could mean that uploading, making a full back-up copy of the human brain would be possible.
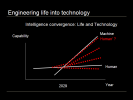
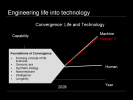
Engineering Life into Technology
Here is a simple graph of the growth in capability of machines and humans over time, machine capacity obviously growing faster than human capacity and the inevitable cross-over point, sometimes called the technological singularity, which could happen, according to futurist Ray Kurzweil, in 2029. We think of life and technology as being disparate, but they need not be, one path forward is to reengineer life into technology that can keep pace with technological advances, shown here as the red Human prime line. In fact, different groups of humanity could adopt different groups of technologies, resulting in more human diversity than we can even imagine. What if Singapore mandated life extension technology adoption and the citizenry became effectively immortal, increasing GDP by one or more orders of magnitude? What if your neighbor’s family decided to have wings? …But the bigger idea I’d like to suggest is that not only is convergence the main path forward but it is already underway…
Status of relevant high-impact research
…as can be seen in key research in the evolving concept of life sciences, genomics, synthetic biology, nanomedicine, and research in intelligence and longevity.
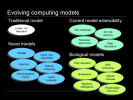
Evolving concept of life sciences
Our concept of life sciences is progressing from an ART to an INFORMATION SCIENCE to now, an ENGINGEERING PROBLEM. The vaunted scientific method is also evolving, supplementing the traditional enumeration and experimentation with two new steps in a virtuous feedback loop, mathematical modeling and computer simulation, and actually building live organisms in the lab to demonstrate mastery.
Our concept of health is also evolving, with desired outcomes upstreaming from cure to improvement, baseline normalization, prevention, enhancement and self-expression; and using a systemic approach, measuring the conditions and symptoms but also looking at genomic data, phenotypic biomarkers and environmental factors. The individual is becoming more of the primary action-taker - measuring, tracking, experimenting, intervening, treating and researching their own conditions, still in consultation with traditional professionals but increasingly in collaboration with peers via health social networks such as Patients Like Me which already has some of the largest aggregated patient communities in the U.S.
The concept of computing is also evolving, as the traditional linear Von Neumann model is extended with new materials, 3D architectures, molecular electronics and solar transistors, and novel models are investigated such as quantum computing, parallel architectures, cloud computing, liquid computing and the cell broadband architecture like that used in the IBM Roadrunner supercomputer. Biological computing models and biology as a substrate are also under exploration: 3D DNA nanotechnology, DNA computing, biosensors, cellular colonies and bacterial intelligence, and the discovery of novel paradigms such as the topological equations by which ciliate DNA is encrypted.

Genomic era Carlson curves: 12 month speedups
This loosening and broadening in the concept of life sciences creates a platform for the genomic era of medicine. In the future, we may look back to a seminal shift between pre-genomic medicine and post-genomic medicine since the genome is the blueprint for understanding how things happen in biology. The killer apps with DNA are sequencing, reading it, and synthesizing, writing it. This chart shows Carlson Curves, the analog to Moore’s Law, a logarithmic plot of the decreasing cost per base pair of DNA sequencing and synthesizing, and indicates quicker speedups, 12 month doublings vs. the 18 month doublings of Moore’s Law. The blue line is DNA sequencing, falling fast recently; the yellow line is base pair synthesizing and the orange line is the synthesizing of short oligo sequences, not dropping as quickly yet and an area of considerable contemporary research.

Direct-to-consumer genomics: 23andme
Not content to wait for institutional medicine to move into the genomic era, consumers have been signing up directly for personalized genomics services. 23andMe is one of the best known companies in the space, and for $399 will sequence 600,000 SNPs and map them to the 80 conditions listed here, ranging from heart disease to ear wax consistency. The individual’s ancestral haplotype group is identified and the raw data can be downloaded, including rsID, chromosome, position and genotype – the As, Cs, Gs and Ts that make you, you! Several people have open sourced their genome by posting it on the web at SNPedia.com

23andme colorectal cancer marker
Drilling down further, this is 23andme’s colorectal cancer marker. The individual’s risk is shown on a scale of 100 and compared to the average. The specific chromosome region, gene and rsID marker, rs6983267 are given along with citations to current scientific research. Heritability vs. environmental factors are discussed, with colorectal cancer being estimated as 35% attributable to genetics. 23andMe is also now including the BRCA1 and 2 markers and a variety of other cancer genes in their service. Direct-to-consumer genomic scanning can be a useful tool for health self-management.

Next-gen sequencing: Pacific Biosciences
But as Carlson Curves suggest, the next generation technology is just around the corner. Pacific Biosciences is one of the leading companies working on high-through put sequencing and projects that their one hour, $100 whole human genome scan, all 3b base pairs, not just 600,000 SNPs, will be available in 2010. The proof of concept paper was published in Jan 2009 describing the company’s technique which is a 30,000-fold improvement on the current method. In step one, the process is to eavesdrop on the body’s natural DNA copying process with the DNA polymerase enzyme, by, in step 2, attaching a fluorescent label to the terminal phosphate of each nucleotide and then in steps 3a and b, detecting and reading the label as it fluoresces and is discarded during nucleotide synthesis using the company’s zero-mode waveguide optical chamber.

Next-gen biotech: synthetic biology
Being able to read and write DNA allows the development of the next generation of biotechnology, synthetic biology; not just altering one gene as with genetic modification but building biological organisms in detail from the bottom up, precisely and reliably.
There are many bioengineering researchers working in the synthetic biology field. Some of the most enthusiastic innovators are the 900 worldwide college students who participated in 2008 international genetically engineered machines competition, iGEM. The winning team from Slovenia created a vaccine for H. pylori ulcers by introducing a synthesized antigen target to bind the H. pylori bacterium’s stealth flagella to the appropriate immune system receptor to which it did not bind to previously. There were several other winners in different categories including a team from Harvard who produced “bactricity,” a bacterial biosensor with electrical output, and a team from IIT Madras who developed a new method to express genes in bacteria using a combination of chemical inputs and heat.
In addition to fundamental research and application, one of the big focus areas in Synthetic Biology is developing tools and technology platforms for engineering biological systems such as those underway at Ginkgo Bioworks.
Another key resource is the Registry of Standard Biological Parts at PartsRegistry.org, an open source DNA parts directory. These off the shelf building blocks encode specific biological functions in snippets of DNA. It is analogous to building a house; its much easier to start with standardized lumber rather than start by growing a tree.
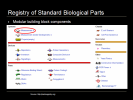
Registry of standard biological parts
Here is the PartsRegistry.org homepage where as of the beginning of 2009, over 3,500 biological parts were listed, organized in three levels of abstraction: parts, devices and systems; all held in a plasmid ready for insertion. Some examples include green fluorescent proteins, cell signaling mechanisms and protein generators and promoters.
For example, clicking on Measurement gives a list of measurement devices.
Parts Registry: select measurement device
Selecting the tetracycline repressible (TetR) Green Fluorescent Protein generator…
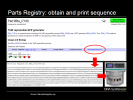
Parts Registry: select measurement device
…brings up the homepage for this function. The PDF datasheet is available and clicking on “Get Selected Sequence” gives the specific DNA snippet for the function which can then be printed into live DNA via a DNA synthesizer or ordered from any one of a few hundred worldwide DNA synthesis labs.
This is the kind of work Craig Venter and others are working on, Venter’s lab has successfully synthesized the full 580,000 base pair (482 genes) genome of mycoplasma genitalium and has now been trying to insert the synthesized genome into an empty nucleus and get the cell to boot.
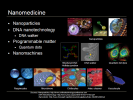
The next important area is nanomedicine which is essentially any health application at the 1-100 nm scale. There is a big focus on nanoparticles and disease treatment due to the targeting capability of the nanoparticle. Cancer nanotechnology is the killer app, so to speak, using nanoparticles made from a variety of materials including gold, calcium phosphate and carbon nanotubes. The nanoparticles may carry a payload, as pictured here in the top row, and be coated on the outside with dye molecules and targeting molecules like folic acid. The particles are injected into the body and travel to and latch onto the cancer cells; this is conceptually straightforward though challenging to execute as cancer cells tends to over-express folic acid receptors. Once the nanoparticles bind with the cancer cells, they may disgorge their payload using nanoporous techniques and be used in conjunction with traditional cancer therapies to improve efficacy as disease cells can be identified and manipulated more precisely.
A second interesting field of nanomedicine is DNA nanotechnology, using DNA as a structural building material rather than as a carrier of genetic information. Structures can be built by forming interlinking strands of DNA into Holliday junctions as pictured here, and then into platforms or tiles. DNA is a useful structural nanotechnology building material particularly because it allows building in 3D, an area where we do not yet have a lot of good tools. DNA nanotechnology is also complementary with other nanotechnologies, for example, carbon nanotubes are stronger and better conductors whereas DNA nanotubes are more easily modified and connected to other structures. One example of a DNA nanostructure is the DNA walker pictured here announced in January 2009 from a lab at Oxford, which in the future, could possibly walk up and down DNA to specific locations to facilitate repair or deliver cargo.
A third area of nanomedicine is programmable matter, matter which has the ability to change its physical properties (shape, density, moduli, optical properties, etc.) in a programmable fashion, based on user input or autonomous sensing. In computing, Intel is looking at programmable matter – creating glass spheres full of electronics that are capable of moving relative to one another changing color and behaving intelligently. Quantum dots are one of the most interesting current health applications of programmable matter. Quantum dots, programmable matter also known as nanocrystals, are essentially mini-semiconductors, 2-10 nanometers (10-50 atoms) in diameter, so small that the addition or removal of an electron changes their properties in some useful way, such as by glowing when stimulated by an external source like ultraviolet (UV) light. Quantum dots are under development as a successor technology to the organic dyes now used ubiquitously in medical research which degrade quickly and have a more limited spectrum of colors, and allow fine structure viewing of cells, pictured here, but being machines, have more biocompatibility and toxicity issues than organic dyes. (A quantum dot is a semiconductor whose excitons are confined in all three spatial dimensions. As a result, they have properties that are between those of bulk semiconductors and those of discrete molecules.)
Fourth is nanomachines, Robert Freitas has designed a variety of species of nanorobots which could eventually supplement or replace several biological functions; respirocytes are artificial RBCs, microbivores are artificial WBS or phagocytes [fag-o-cytes], clottocytes are artificial platelets and there are a number of artery cleaners and vasculocytes for artery repair. These nanomachines require a substantial amount of engineering and are at minimum 15-20 years out since we have yet to make any real progress in mechanosynthesis, the underlying techniques of adding and removing atoms in molecular nanotechnology, building atomically precise matter from the bottom up. (Diamond mechanosynthesis is mechanically adding and removing Hydrogen atoms and depositing Carbon atoms.

Radio-surgery and on-demand physicians
There are many interesting advances in applied medicine. OR-Live.com is providing live webcasts from operating rooms. It’s a great tool for patients to review a potential procedure and for physicians to collaborate and train. Questions can be emailed in live to the operating team who may respond during the webcast. OR-Live is also a good way to see new technologies in use, such as the da Vinci robotic surgery platform which makes procedures much less invasive with small incisions or ports, so that thin tubes with the robotic arms can be inserted into the patient’s body. Then the physician performs the surgery from a computer terminal, manipulating the robotic arms.
Another surgical platform, not even requiring a physician in the treatment room is Accuray’s cyberknife radiosurgery where linear-accelerator generated radiation vectors treat tumors, even in the brain as 3D cams adjust in real-time to patient breathing and other movements.
Telemedicine is also growing, with X-rays and other applications such as on-demand physician consultation using web cams and instant messenger-style chat for the price of a standard co-pay as offered by American Well.com.

Simulation, modeling and virtual health
Simulation and virtual worlds are emerging as important life sciences tools. Entelos and Optimata are two companies with virtual patient biosimulation technology. Predictive biosimulation is using empirical biological data and mathematical modeling to create an electronic version of an individual, a virtual patient on which to test simulated treatments. An example is pictured here illustrating the baseline case (x-axis) as compared to the expected improvement from a potential intervention (diagonal line). In December 2008, the U.S. FDA announced plans to use the Entelos biosimulation technology to study three heart-related drugs to identify safety and effectiveness issues before the completion of late-stage human trials by testing far more simulated patients than could be included conventionally and obtaining computer-generated test results in days or weeks instead of years.
Next are some examples of the open source BioSPICE biosimulation platform and virtual worlds, providing a mind’s eye view of simulating biological processes, here from the point of view of being inside the testis, and two virtual hospitals allowing patient orientation and physician training and collaboration.

Intelligence research by approach
After the mechanics of working with DNA and other biological processes, the next obvious area to address for the convergence of life and technology is the nervous system, particularly the brain. There are many different approaches to brain research ranging from understanding the examples we have to simulating and building intelligence.
In computer simulation and whole brain emulation, the leading effort is the Blue Brain project led by Henry Markram at the Ecole Polytechnique Federale in Lausanne, Switzerland. The project achieved a critical milestone in 2007 by building an accurate computer-based replica of one neocortical column of a rat brain. A key finding of the team is that the neocortical column, or NCC, is the critical and modular component of the brain, having between 10,000 and 100,000 neurons depending on the species and neocortical region; then that millions of neocortical columns comprise the full brain. In November 2008, IBM reported that the current progress of the project is emulating the full cortex of a 2-month old rat at one-tenth real-time speed on a dedicated IBM Blue Gene L supercomputer. The team estimates that it will take another ten years, until 2018, to emulate a full human brain at real-time speed.
In October 2008, Oxford University’s Nick Bostrom and Anders Sandberg published the Whole Brain Emulation Roadmap, a detailed estimate of the key steps and progression necessary for the whole brain emulation of a human.
As diskdrive and battery technology advances enabled the development of the iPod, technology progress in adjacent fields has been critical to whole brain emulation, particularly neuroimaging, infrared-DIC (differential interference control), videomicroscopy to record actual synapse activity and complex neuronal simulation software.
The second area, neuroimaging of tissue volumes, has also been progressing. The leading labs at Harvard (Jeff Lichtman) and Texas A&M (Yoonsuck Choe) have demonstrated synapse and vesicle detection in the 300nm wide brain tissue slices generated and examined with their KESM (knife-edge scanning microscopy) methodology. The goal of these groups is connectomics, creating a circuit diagram of the brain by mapping every synapse and neural connection.
The third important area in intelligence research is integrating life and technology via brain-computer interfaces, also known as cognitive enhancement or neuroengineering, and body area networks. The goal of brain-computer interfaces is enhancing human brain function by establishing bi-directional communication between the mind and external storage devices, initially in the areas of memory and learning. The most promising progress to date is with vision systems. In fact, 10% of Americans are already cyborgs in the sense of having permanently implanted inorganic devices in their bodies such as pacemakers, hips and teeth.
Body-area networks right now are basic; consisting of one or a few implanted or wearable biosensors collecting basic biological data and transmitting it wirelessly to a computer. The next phase could include larger more complex networks of intercommunicating sensors and eventually sensors empowered to take action or otherwise behave autonomously, and two-way broadcast body-area networks, with the ability to bring communications on-board, eventually possibly being your own wifi hotspot. The IEEE is working on body-area networking standards as protocol 802.15.6.
One of the biggest challenges with devices implanted in the body is energy; providing adequate ongoing power to the device. Power trumps the other two concerns: bandwidth and biocompatibility. Many interesting methods of power generation are being investigated including thermal and vibrational energy, RF, PV, bio-chemical energy, using elastic DNA energy to move a particle along the DNA, and the ATP chip, possibly getting nanodevices to produce ATP from naturally circulating glucose.
Robotics and artificial intelligence are the fourth area of intelligence research showing advances with robots for military applications like the Boston Dynamics’ Big Dog pack robot and at the other end of the spectrum, Leonardo from Cynthia Brazeal’s lab at MIT which is more empathic than many humans. Current funding for narrow or specific AI applications comes from DARPA for projects such as the Grand Challenge unmanned urban navigation vehicle, and corporate research budgets for projects such as expert systems that examine seismographic data. Strong AI for general purpose problem solving is the focus of a few innovative startups and there is considerable debate about a variety of issues, including how to create a moral or friendly AI.

Longevity: extending lifespan and healthspan
Once all biological processes are understood and manageable including disease and cure, there is no reason why a human’s lifespan and healthspan could not be indefinite.
The leader in addressing aging as a pathology that should be cured is Aubrey de Grey, a British biomedical gerontologist. He recognizes that aging is highly multidisciplinary, and brings the fields of cancer, stem cells, immunology, tissue generation, genetic therapy and regenerative medicine together under one umbrella.
In de Grey’s Strategies for Engineered Negligible Senescence (SENS) program, aging is distilled into 7 problems to tackle:
- DNA mutations in the cell nucleus and mitochondria
- Junk that builds up inside and outside the cells
- Cell loss and death, and
- Extra-cellular crosslinks, when cells become stuck together
De Grey outlines potential solutions to each of these problems and has funded several scientists through his SENS research foundation. Possible solutions range from the incremental to the disruptive, here are three of the most disruptive ideas. First is adapting enzymes used in environmental remediation, from microbes that eat oil spills, and investigating inserting them into the body’s lysosomes to eat amyloid plaques that can lead to Alzheimer’s disease and atherosclerotic plaques that can lead to heart disease; some of these LysoSENS cells are pictured here. A second idea is transferring the relevant genes from the mitochondrial DNA to the nucleus where they are better protected from mutation. A third even more radical idea is 100% cancer prevention by deleting the telomerase gene, essentially prohibiting all cells from the out-of-control divisions of cancer; this would require stem cell therapies each 5-10 years so that normal cell division could continue.
Finally, an important concept is escape velocity, the idea of living healthfully long enough for these therapies to start to become available.
Without these or other changes, we are on a slow and linear improvement curve for life span, expecting less growth in the next 50 years than was realized in the last 50 years; 8% vs. 12% previously. You can evaluate your own real age vs. biological age as a function of lifestyle and other factors at RealAge.com.
…So, after seeing these research updates, what are the implications for the future?
Engineering life into technology
The research described here creates the foundations of convergence. The expanding concepts of life sciences, health and computing, the advent of the genomic era of medicine, synthetic biology, building organisms from the bottom up as the next-generation of biotechnology, nanomedicine as a more precise way of working with biology, intelligence research providing a deeper understanding of the brain and longevity research working to remedy the pathology of aging.

What can be expected in the short-term?
What kinds of signals of convergence can we look for in the next several years?
One key trend that seems likely to continue is the miniaturization of electronics. In 1970, a computer was the size of a room, today a smart cell phone has the same processing power. In another 15-20 years computers might be invisible to the unaugmented human eye, easily wearable or implantable.
A second occurrence could be that whole human genome scans become as routine as blood tests, that everyone has one on file with their doctor. Huge databases of anonymized whole human genomes could facilitate clinical trials with patient groups pre-aggregated and pre-qualified substantially accelerating the era of pharmacogenomics and personalized medicine where every treatment is tailored to the individual.
Third, a new generation of biotech including synthetic biology and nanomedicine could follow cleantech as the next powerhouse of entrepreneurial development and VC financing, particularly with a focus on key cures in areas like obesity and cancer.
Fourth, peer-to-peer health, now where peer-to-peer finance was 5 years ago, could blossom as more people engage with health social networks for any variety of health issues, not just disease cure. The long-tail of medicine could be enabled as patients with so-called orphan conditions are aggregated into identifiable online patient groups that can approach and be approached for research and solutions.
Fifth, a culture of biohacking could spring up, mirroring infotech hacking culture, as the new definition of literacy, creativity, opportunity and fun may include being able to create biological organisms.

Ultimate possibilities for life and technology
Thinking longer-term, what would it be like if all matter, including life, could be designed to spec with nanotechnology and synthetic biology? Form factor could become ephemeral and purpose-driven. An intelligence could embody as a human, as a fleet of starships, as a crane, as a school of nanoparticles, or remain digital.
Some interesting issues could come up, say from having multiple persistent copies of one intelligence. What would the social, legal, economic etiquette and governing laws be? Or would these words even make sense anymore? Will the notion of the distinct individual become obsolete?
What about transhumanism, when groups or all humans have radically different capabilities than today? Or posthumanism, the moment of speciation?
What about utility functions? In a digital format, traditional biological functions make a lot less sense. And what about emotion? Is there a relevant adaptation for the digital substrate or is emotion just another biology-based information system?
What is intelligence and is it reflected differently in a digital medium without the sensory input context of the physical world? Maybe intelligence is nothing more than manipulating patterns of information.
Slides: http://www.slideshare.net/lablogga/future-of-life-sciences-1240003